I am working on an update of simstudy that will make generating survival/time-to-event data a bit more flexible. There are two biggish enhancements. The first facilitates generation of competing events, and the second allows for the possibility of generating survival data that has time-dependent hazard ratios. This post focuses on the first enhancement, and a follow up will provide examples of the second. (If you want to try this at home, you will need the development version of simstudy, which you can install using devtools::install_github(“kgoldfeld/simstudy”).)
Competing risks
In standard survival analysis, we are interested in understanding the distribution of time to a single event, such as time to death or time to symptom-free recovery. However, there may be situations where two there are at least two possible events, and the occurrence of one precludes the occurrence of the other. In this case, we say that the events are competing risks. In the work that motivated this simstudy enhancement, we are trying to model the time to opioid overdose death for people being released from jail, but there are at least two other competing risks, death from other causes and re-incarceration. In addition, there is censoring, which from a data generation point of view (though maybe not from an analytic perspective), can also be considered a competing risk.
Generating competing risk outcomes under 0.4.0
Previously, it was possible to generate competing risk outcomes, but it had to be done “manually” with additional lines of code or data definitions. It was a bit cumbersome:
library(simstudy)
library(data.table)
library(survival)First, we would generate a data set with genSurv:
d1 <- defData(varname = "x1", formula = .5, dist = "binary")
d1 <- defData(d1, "x2", .5, dist = "binary")
dS <- defSurv(varname = "event_1", formula = "-12 - 0.1*x1 - 0.2*x2", shape = 0.3)
dS <- defSurv(dS, "event_2", "-12 - 0.3*x1 - 0.2*x2", shape = 0.3)
dS <- defSurv(dS, "event_3", "-12 - 0.4*x1 - 0.3*x2", shape = 0.3)
dS <- defSurv(dS, "censor", "-13", shape = 0.3)
set.seed(2140)
dtCov <- genData(3001, d1)
dtSurv <- genSurv(dtCov, dS)
head(dtSurv)## id x1 x2 censor event_1 event_2 event_3
## 1: 1 0 1 56.809 40.321 35.050 48.356
## 2: 2 0 0 62.278 24.865 23.121 19.198
## 3: 3 1 0 25.650 27.898 24.716 49.633
## 4: 4 1 1 51.486 35.065 44.731 40.086
## 5: 5 1 0 54.522 37.042 49.701 34.751
## 6: 6 1 0 42.749 41.419 34.136 43.811And then we would add the observed time variable with a new data definition and call to addColumns:
f <- "(time==censor)*0 + (time==event_1)*1 + (time==event_2)*2 + (time==event_3)*3"
cdef <- defDataAdd(varname = "time",
formula = "pmin(censor, event_1, event_2, event_3)", dist = "nonrandom")
cdef <- defDataAdd(cdef, varname = "event",
formula = f,
dist = "nonrandom")
dtSurv_final <- addColumns(cdef, dtSurv)
head(dtSurv_final)## id x1 x2 censor event_1 event_2 event_3 time event
## 1: 1 0 1 56.809 40.321 35.050 48.356 35.050 2
## 2: 2 0 0 62.278 24.865 23.121 19.198 19.198 3
## 3: 3 1 0 25.650 27.898 24.716 49.633 24.716 2
## 4: 4 1 1 51.486 35.065 44.731 40.086 35.065 1
## 5: 5 1 0 54.522 37.042 49.701 34.751 34.751 3
## 6: 6 1 0 42.749 41.419 34.136 43.811 34.136 2Streamlined approach
The function addCompRisk can now generate an observed time variable (which will be the first of the four event times). This is done by specifying a timeName argument that will represent the observed time value. The event status is indicated in the field set by the eventName argument (which defaults to “event”). And, if a variable name is indicated in the censorName argument, the censored events automatically have a value of 0.
dtSurv_final <- addCompRisk(dtSurv,
events = c("event_1", "event_2", "event_3", "censor"),
timeName = "time", censorName = "censor")
head(dtSurv_final)## id x1 x2 time event type
## 1: 1 0 1 35.050 2 event_2
## 2: 2 0 0 19.198 3 event_3
## 3: 3 1 0 24.716 2 event_2
## 4: 4 1 1 35.065 1 event_1
## 5: 5 1 0 34.751 3 event_3
## 6: 6 1 0 34.136 2 event_2The competing risk data can be plotted using the cumulative incidence functions (rather than the survival curves):
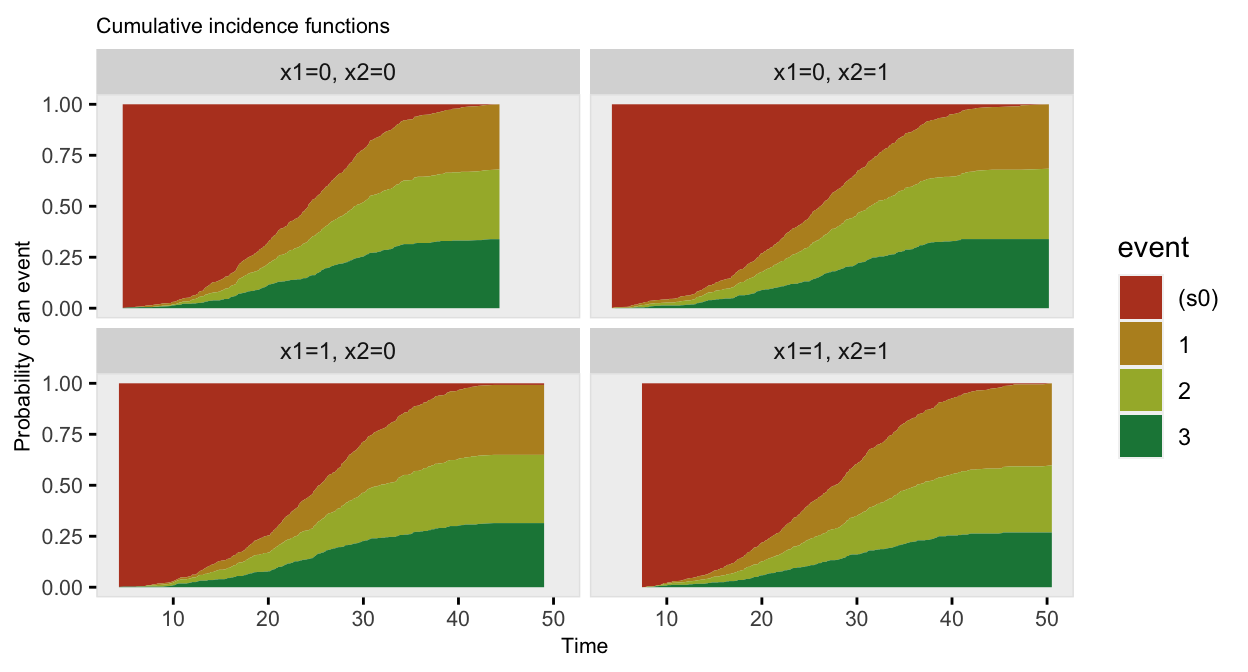
The data generation can be streamlined even further, as there is no need to call addCompRisk at all. The same arguments can be provided directly in the call to genSurv, so everything is done at once. And there is no need to explicitly name the events, as they are extracted from the survival distribution definitions:
dtSurv_final <- genSurv(dtCov, dS, timeName = "time", censorName = "censor")
head(dtSurv_final)## id x1 x2 time event type
## 1: 1 0 1 30.229 3 event_3
## 2: 2 0 0 28.473 1 event_1
## 3: 3 1 0 11.654 0 censor
## 4: 4 1 1 28.248 1 event_1
## 5: 5 1 0 28.524 3 event_3
## 6: 6 1 0 18.255 0 censorIf you don’t want to generate competing risk data, and you can set timeName to NULL, or just not specify it:
dtSurv_final <- genSurv(dtCov, dS)
head(dtSurv_final)## id x1 x2 censor event_1 event_2 event_3
## 1: 1 0 1 40.276 16.858 42.624 34.609
## 2: 2 0 0 69.382 24.191 37.574 10.783
## 3: 3 1 0 26.637 48.476 54.240 60.507
## 4: 4 1 1 36.697 46.690 36.443 43.195
## 5: 5 1 0 64.498 36.755 39.666 10.935
## 6: 6 1 0 69.150 38.706 44.930 17.668