On an incredibly heartening note, two COVID-19 vaccines have been approved for use in the US and other countries around the world. More are possibly on the way. The big challenge, at least here in the United States, is to convince people that these vaccines are safe and effective; we need people to get vaccinated as soon as they are able to slow the spread of this disease. I for one will not hesitate for a moment to get a shot when I have the opportunity, though I don’t think biostatisticians are too high on the priority list.
Those who are at the top of the priority list are staff and residents of nursing homes. Unfortunately, within these communities, there are pockets of ambivalence or skepticism about vaccines in general, and the COVID vaccine in particular. In the past, influenza vaccine rates for residents at some facilities could be as low as 50%. I am part of an effort organized by researchers affiliated with the IMPACT Collaboratory to figure out a way to increase these numbers.
This effort involves a cluster randomized trial (CRT) to evaluate the effectiveness of a wide-ranging outreach program designed to encourage nursing home residents to get their shots. The focus is on facilities that have relatively high proportions of African-American and Latinx residents, because these facilities have been among the least successful in the past in convincing residents to get vaccinated. The outcome measure of the trial, which will be measured at the individual level, will be the probability of vaccination within 5 weeks of being available at the nursing home.
The nursing homes in the study are members of one of four national nursing home networks or corporations. In this CRT, the randomization will be stratified by these networks and by the proportion of African-American and Latinx residents. We are defining the race/ethnicity strata using cutoffs of proportions: <25%, 25% to 40%, and >40%. We want our randomization to provide balance with respect to racial/ethnic composition in the intervention and control arms within each individual stratum. However, we are concerned that the strata with fewer nursing homes have a high risk of imbalance just by chance. Constrained randomization is one possible way to mitigate this risk, which is the focus here in this post.
Constrained randomization
The basic idea is pretty simple. We generate a large number of possible randomization lists based on the requirements of the study design. For each randomization, we evaluate whether the balancing criteria have been met; by selecting only the subset of randomizations that pass this test, we create a sample of eligible randomizations. With this list of possible randomizations is in hand, we randomly select one, which becomes the actual randomization. Because we have limited the final selection only to possible randomizations that have been vetted for balance (or whatever criteria we require), we are guaranteed to satisfy the pre-specified criteria.
Simulated data
I am using a single simulated data set to illustrate the constrained randomization process. Using the simstudy package, creating this data set is a two-step process of defining the data and then generating the data.
Defining the data
There will be a total of 200 nursing homes in 3 (rather than 4) networks. Just as in the real data, racial/ethnic composition will differ by network (because they are based in different parts of the country). And the networks are different sizes. The proportions of African-American/Latinx residents are generated using the beta distribution, which ranges from 0 to 1. In simstudy, the beta distribution is parameterized with a mean (specified in the formula argument) and dispersion (specified in the variance argument. See this for more details on the beta distribution.
library(simstudy)
library(data.table)
def <- defData(varname = "network", formula = "0.3;0.5;0.2",
dist = "categorical", id = "site")
defC <- defCondition(condition = "network == 1",
formula = "0.25", variance = 10, dist = "beta")
defC <- defCondition(defC, condition = "network == 2",
formula = "0.3", variance = 7.5, dist = "beta")
defC <- defCondition(defC, condition = "network == 3",
formula = "0.35", variance = 5, dist = "beta")Generating the data
set.seed(2323761)
dd <- genData(200, def, id = "site")
dd <- addCondition(defC, dd, newvar = "prop")
dd[, stratum := cut(prop, breaks = c(0, .25, .4, 1),
include.lowest = TRUE, labels = c(1, 2, 3))]
dd## site prop network stratum
## 1: 1 0.340 2 2
## 2: 2 0.181 2 1
## 3: 3 0.163 2 1
## 4: 4 0.099 3 1
## 5: 5 0.178 2 1
## ---
## 196: 196 0.500 2 3
## 197: 197 0.080 3 1
## 198: 198 0.479 3 3
## 199: 199 0.071 2 1
## 200: 200 0.428 2 3Randomization
Now that we have a data set in hand, we can go ahead an randomize. I am using the simstudy function trtAssign, which allows us to specify the strata as well as the the ratio of controls to intervention facilities. In this case, we have a limit in the number of sites at which we can implement the intervention. In this simulation, I assume that we’ll randomize 150 sites to control, and 50 to the intervention, a 3:1 ratio.
dr <- trtAssign(dd, nTrt = 2, balanced = TRUE, strata = c("network", "stratum"),
ratio = c(3, 1), grpName = "rx")We want to inspect an average proportion of African-American/Latinx residents within each strata (without adjusting for nursing home size, which is ignored here). First, we create a data table that includes the difference in average proportions between the facilities randomized to the intervention and those randomized to control:
dx <- dr[, .(mu_prop = mean(prop)), keyby = c("network", "stratum", "rx")]
dc <- dcast(dx, network + stratum ~ rx, value.var = "mu_prop")
dc[, dif := abs(`1` - `0`)]Looking at both the table and the figure, Stratum 3 (>40%) in Network 1 jumps out as having the largest discrepancy, about 15 percentage points:
## network stratum 0 1 dif
## 1: 1 1 0.16 0.192 0.03062
## 2: 1 2 0.31 0.305 0.00641
## 3: 1 3 0.45 0.600 0.14568
## 4: 2 1 0.15 0.143 0.00394
## 5: 2 2 0.32 0.321 0.00310
## 6: 2 3 0.49 0.547 0.05738
## 7: 3 1 0.13 0.085 0.04948
## 8: 3 2 0.33 0.331 0.00029
## 9: 3 3 0.56 0.607 0.04284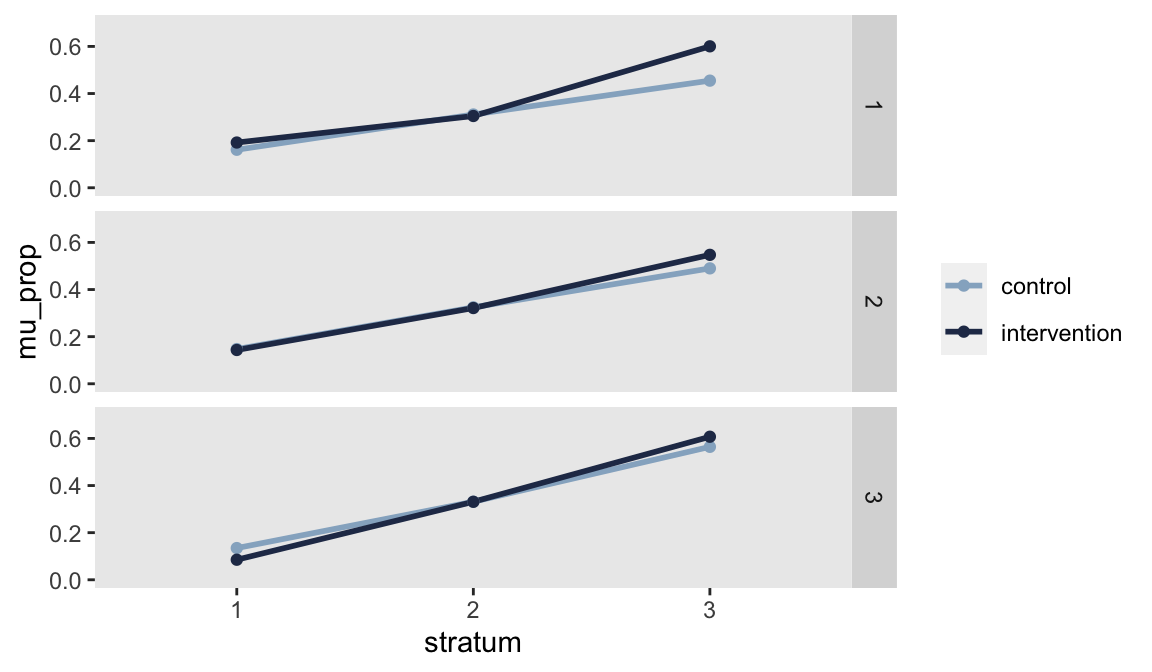
Constraining the randomization
We want to do better and ensure that the maximum difference within a stratum falls below some specified threshold, say a 3 percentage point difference. All we need to do is repeatedly randomize and then check balance. I’ve written a function randomize that will be called repeatedly. Here I generate 1000 randomization lists, but in some cases I might need to generate many more, particularly if it is difficult to achieve targeted balance in any particular randomization.
randomize <- function(dd) {
dr <- trtAssign(dd, nTrt = 2, strata = c("network", "stratum"), balanced = TRUE,
ratio = c(3, 1), grpName = "rx")
dx <- dr[, .(mu_prop = mean(prop)), keyby = c("network", "stratum", "rx")]
dx <- dcast(dx, network + stratum ~ rx, value.var = "mu_prop")
dx[, dif := abs(`1` - `0`)]
list(is_candidate = all(dx$dif < 0.03), randomization = dr[,.(site, rx)],
balance = dx)
}
rand_list <- lapply(1:1000, function(x) randomize(dd))Here is one randomization that fails to meet the criteria as 5 of the 9 strata exceed the 3 percentage point threshold:
## [[1]]
## [[1]]$is_candidate
## [1] FALSE
##
## [[1]]$randomization
## site rx
## 1: 1 0
## 2: 2 1
## 3: 3 0
## 4: 4 1
## 5: 5 0
## ---
## 196: 196 0
## 197: 197 1
## 198: 198 0
## 199: 199 0
## 200: 200 0
##
## [[1]]$balance
## network stratum 0 1 dif
## 1: 1 1 0.16 0.207 0.0503
## 2: 1 2 0.30 0.334 0.0330
## 3: 1 3 0.45 0.600 0.1457
## 4: 2 1 0.15 0.138 0.0107
## 5: 2 2 0.32 0.330 0.0078
## 6: 2 3 0.50 0.514 0.0142
## 7: 3 1 0.13 0.085 0.0493
## 8: 3 2 0.34 0.311 0.0239
## 9: 3 3 0.55 0.647 0.0950Here is another that passes, as all differences are below the 3 percentage point threshold:
## [[1]]
## [[1]]$is_candidate
## [1] TRUE
##
## [[1]]$randomization
## site rx
## 1: 1 1
## 2: 2 0
## 3: 3 1
## 4: 4 0
## 5: 5 1
## ---
## 196: 196 1
## 197: 197 0
## 198: 198 1
## 199: 199 1
## 200: 200 0
##
## [[1]]$balance
## network stratum 0 1 dif
## 1: 1 1 0.16 0.18 0.0168
## 2: 1 2 0.31 0.31 0.0039
## 3: 1 3 0.49 0.49 0.0041
## 4: 2 1 0.15 0.14 0.0144
## 5: 2 2 0.32 0.33 0.0064
## 6: 2 3 0.50 0.52 0.0196
## 7: 3 1 0.12 0.12 0.0095
## 8: 3 2 0.34 0.31 0.0239
## 9: 3 3 0.57 0.58 0.0134All that remains is to identify all the randomization sets that met the criteria (in this case there are only 6, suggesting we should probably generate at least 100,000 randomizations to ensure we have enough to pick from).
candidate_indices <- sapply(rand_list, function(x) x[["is_candidate"]])
candidates <- rand_list[candidate_indices]
(n_candidates <- length(candidates))## [1] 6selected <- sample(x = n_candidates, size = 1)
ds <- candidates[[selected]][["randomization"]]
ds <- merge(dd, ds, by = "site")
dx <- ds[, .(mu_prop = mean(prop)), keyby = c("network", "stratum", "rx")]And looking at the plot confirms that we have a randomization scheme that is balanced based on our target:
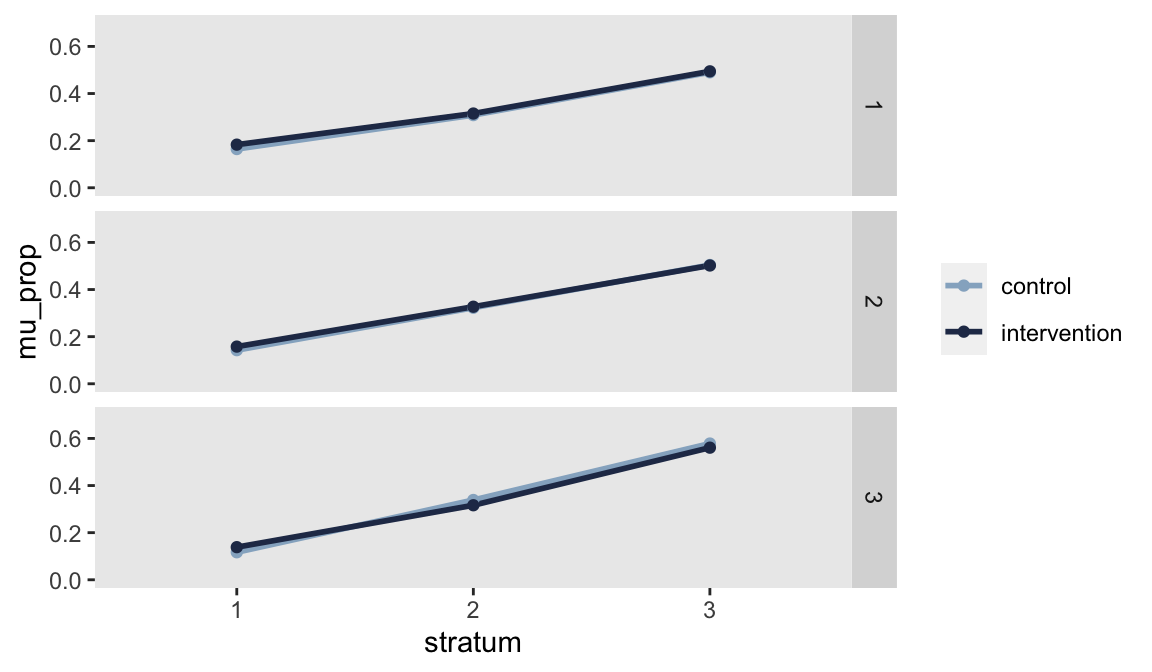
Of course, the selection criteria could be based on any combination of factors. We may have multiple means that we want to balance, or we might want the two arms to be similar with respect to the standard deviation of a measure. These additional criteria may require more randomization schemes to be generated just because balance is that much more difficult to achieve, but all that really costs is computing time, not programming effort.
Support:
This work was supported in part by the National Institute on Aging (NIA) of the National Institutes of Health under Award Number U54AG063546, which funds the NIA IMbedded Pragmatic Alzheimer’s Disease and AD-Related Dementias Clinical Trials Collaboratory (NIA IMPACT Collaboratory). The author, a member of the Design and Statistics Core, was the sole writer of this blog post and has no conflicts. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.